Antibodies stand as the leading class of protein therapeutics, with over 160 licensed antibody therapeutics across the globe. The market for these innovative therapies is projected to soar, reaching an astounding US$445 billion within the next five years. The journey of antibody development typically unfolds in two essential phases: the discovery of antibodies that specifically bind to targeted epitopes, followed by the process of affinity maturation and clinical optimization of these antibodies.
The current methodologies for identifying epitope-specific antibodies predominantly depend on animal immunization or the screening of antibody libraries. These processes aim to pinpoint candidate molecules capable of binding to desired targets, followed by extensive epitope mapping. However, these traditional methods are often laborious, time-consuming, and may fail to uncover antibodies that interact with therapeutically relevant epitopes.
Many endeavors in the computational design of antibodies have concentrated on the optimization phase, which includes sampling alternative native CDR loops to enhance affinities or leveraging Rosetta sequence design techniques to refine interacting regions. Recent innovations have incorporated structure-based and sequence-based deep learning networks trained to create novel antibody sequence variants. Yet, these methods necessitate an existing binding antibody for optimization. There have also been strides in antibody optimization through deep learning methods utilizing data generated from new experimental techniques.
Despite these advancements, there remains a critical gap: computational methods capable of executing the initial stage of antibody design—generating epitope-specific binding antibodies—are virtually non-existent. The challenge of de novo antibody design, which does not rely on homology to existing antibodies targeting the same epitope, remains unsolved. While there has been rapid progress in binding protein design using RFdiffusion, these methods predominantly depend on regular secondary structure-based interactions with target epitopes.
We hypothesized that a specialized version of RFdiffusion, fine-tuned specifically on antibody structures, could effectively design de novo CDR-mediated interfaces. This hypothesis is grounded in the understanding that the underlying thermodynamics of interface formation remain consistent, regardless of the specific target. Our aim was to develop a method that enables the targeting of any specified epitope on any desired target, focuses sampling on CDR loops, and explores alternative rigid-body placements of the designed antibody relative to the epitope.
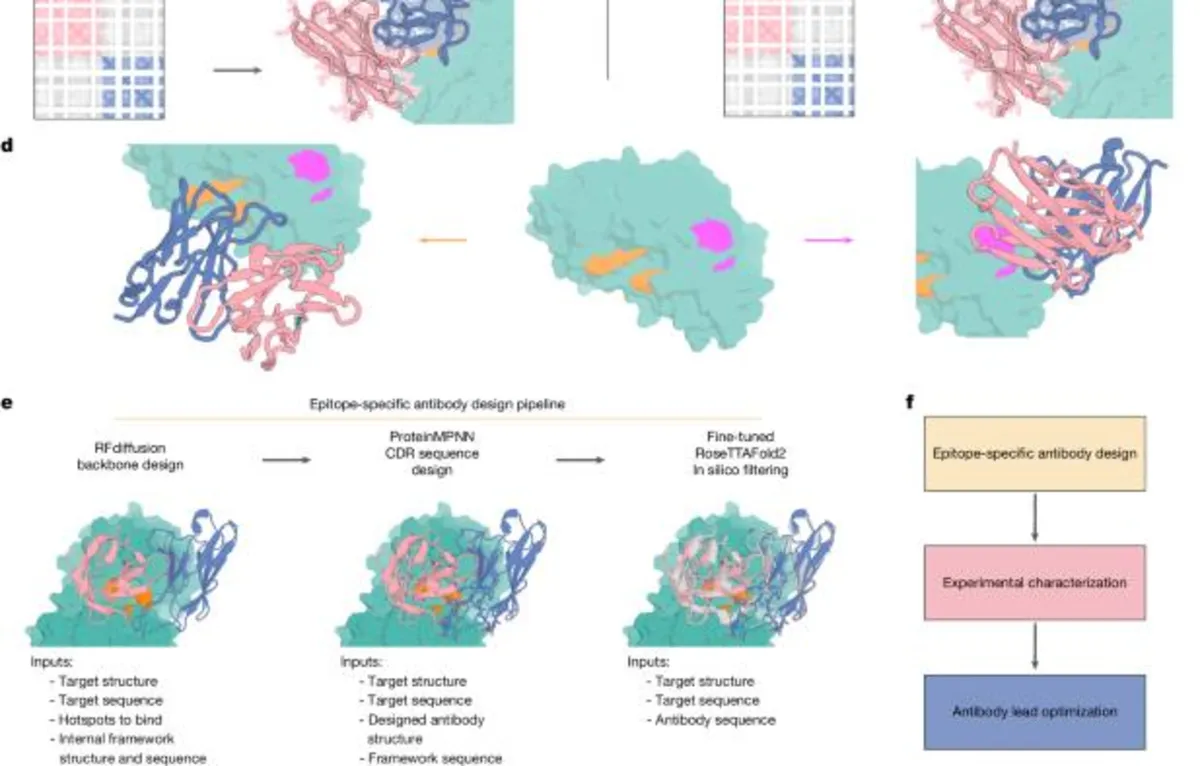
RFdiffusion employs the AlphaFold2 framework along with RF2 representations of protein backbones. During the training phase, a noising schedule is executed over a set number of timesteps to corrupt protein frames towards random prior distributions. The training involves sampling a Protein Data Bank (PDB) structure and applying noising steps, with the aim of minimizing mean squared error loss between true and predicted structures. We fine-tuned RFdiffusion primarily using antibody complex structures, allowing for the specification of the framework structure and sequence during inference.
Through this innovative training regimen, RFdiffusion has demonstrated the ability to design antibody structures that closely align with specified framework structures while targeting designated epitopes with novel CDR loops. Subsequent to the RFdiffusion phase, we utilized ProteinMPNN to design the sequences of the CDR loops, resulting in antibodies that exhibit diverse interactions with target epitopes, significantly different from the training dataset.
To ensure the experimental success of our designs, we implemented effective filtering methods based on the similarity of designed structures to AlphaFold2-predicted structures. This strategy aims to select the proteins and interfaces most likely to succeed experimentally, which is particularly challenging in the case of antibodies, as AlphaFold2 has limitations in accurately predicting antibody-antigen structures. To enhance our design filtering capabilities, we fine-tuned RoseTTAFold2 on antibody structures, incorporating information about target structure and epitope location.
Initially, our focus was on designing single-domain antibodies (VHHs) derived from camelids, which have shown significant promise in therapeutic applications. Two VHH therapies have already received FDA approval, with numerous clinical trials ongoing. Despite possessing fewer CDR loops than conventional antibodies, VHHs present a comparable average interaction surface area, suggesting that methodologies effective for VHH design could also apply to broader antibody design.
We designed VHHs targeting various disease-relevant targets, including C. difficile TcdB and the SARS-CoV-2 receptor-binding domain. The computationally filtered designs underwent high-throughput screening via yeast surface display or lower-throughput screening with Escherichia coli expression and surface plasmon resonance. Our results revealed high-affinity binders with distinct CDR loops, showcasing the capability of RFdiffusion to create antibodies that engage specific epitopes.
To assess the accuracy of our designs, we employed cryo-electron microscopy (cryo-EM) to determine the structures of designed VHHs in complex with target proteins. Our findings confirmed that designed VHHs effectively bound to the trimeric influenza haemagglutinin glycoprotein, showcasing remarkable alignment with predicted models and validating our computational design approach.
To enhance the binding affinity of our de novo-designed VHHs, we utilized the OrthoRep system for continuous hypermutation. This approach has been proven to accelerate the affinity maturation of antibodies. The affinity-matured VHHs achieved significant improvements in binding affinities, making them viable candidates for downstream cryo-EM structural characterization.
While our results establish the feasibility of de novo antibody design, the experimental success rates indicate room for improvement. The incorporation of advanced filtering methods and the refinement of training protocols can further enhance the efficacy of our designs. As computational methods in antibody design advance, they hold the potential to revolutionize the discovery and development of antibodies, paving the way for new therapeutic applications.
The computational design of antibodies using RFdiffusion and related techniques opens new avenues for targeting specific epitopes on therapeutic proteins. As we continue to refine these methods and improve experimental success rates, we expect a significant expansion in the range of clinical targets and diseases amenable to antibody therapeutics.