The Democratic Republic of the Congo (DRC) is currently grappling with multiple health crises, including outbreaks of mpox, cholera, and malaria. On September 4, 2025, the Ministry of Public Health in the DRC officially declared the 16th outbreak of Ebola Virus Disease (EVD) after confirming the presence of the Ebola virus (EBOV), formerly known as Zaïre ebolavirus, in patient specimens from the Bulape Health Zone in Kasai Province.
The resurgence of EBOV in remote areas with limited healthcare access is not unprecedented; the DRC has experienced 15 previous Ebola outbreaks. In late August 2025, local health authorities in Bulape raised alarms over cases exhibiting symptoms of haemorrhagic fever and associated fatalities among both patients and healthcare workers. In response, health officials collected a buccal swab from a fatal case and five blood specimens from six suspected cases, which were sent to the Institut National de Recherche Biomédicale (INRB) in Kinshasa for confirmation and whole genome sequencing (WGS).
On September 3, 2025, the INRB conducted tests on the six samples using the point-of-care GeneXpert Ebola assay, the automated BioFire FilmArray System, and real-time PCR with the Altona RealStar Filovirus RT-PCR kit. Samples that tested positive for EBOV were sequenced using a nanopore sequencing device from Oxford Nanopore Technology. The process began with viral RNA extraction from 140 µL of inactivated specimens using a QIAGEN viral RNA extraction kit, following the manufacturer's protocol.
Subsequently, reverse transcription was performed using the LunaScript RT SuperMix kit from New England Biolabs. A multiplex PCR was conducted on the complementary DNA (cDNA) with the Q5 Hot Start High-Fidelity 2X master mix, applying EBOV-specific primer pools designed for EBOV Mangina variants to amplify the viral genome. The sequencing library was prepared in triplicate using the rapid sequencing DNA V14 barcoding kit from Oxford Nanopore Technologies, and sequenced on a GridION sequencer. The reads were subsequently base-called with the High accuracy model from Guppy.
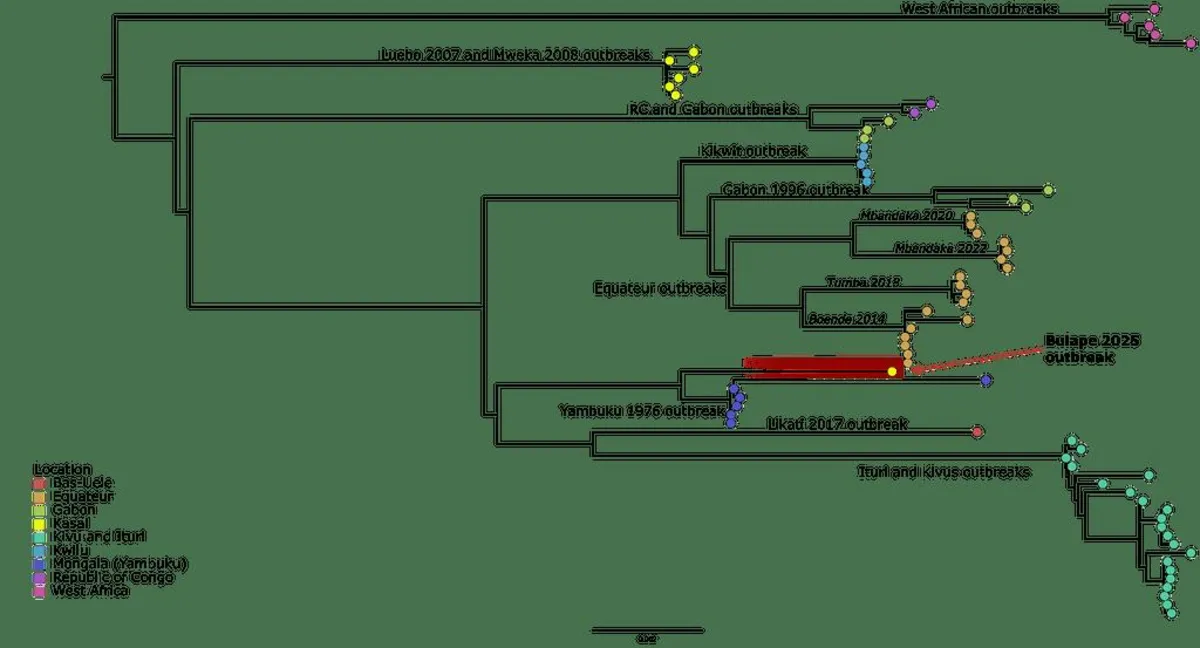
In less than an hour, a highly accurate consensus genome was generated, demonstrating that the new EBOV genome is 99.97% complete and comprises 18,819 nucleotides. The analysis included a multiple sequence alignment of 71 complete EBOV genomes, including the newly generated genome, executed using MAFFT. A phylogenetic tree was constructed with IQ-TREE v2.1.4, revealing that the new EBOV genome has a nucleotide similarity of 99.52% to the nearest related genome, Ebola virus/H.sapiens-tc/COD/1976/Yambuku-Mayinga (GenBank accession number NC_002549.1). This finding suggests that the current outbreak represents a new zoonotic spillover event rather than a continuation of the 2007 Luebo or 2008/2009 Mweka EVD outbreaks.
The study involved key contributors in various roles, including data collection, molecular testing, data interpretation, WGS, bioinformatics analysis, and manuscript writing. Notable contributors include:
Adrienne Amuri-Aziza Gradi Luakanda-Ndelemo Jean-Claude Makangara-Cigolo Prince Akil-Bandali Servet Kimbonza Fiston Cikaya-Kankolongo Princesse Paku-Tshambu Elzedek Mabika-Bope Emmanuel Lokilo André Citenga Raphael Lumembe Gabriel Kabamba Christian Ngandu Louis Tshulo Mathias Mossoko Dieudonné Mwamba Elisabeth Pukuta Eddy Kinganda-Lusamaki Patrick Mukadi Dieudonné Mumba Ngoyi Steve Ahuka-Mundeke Jean-Jacques Muyembe-Tamfum Tony Wawina-Bokalanga Placide Mbala-KingebeniWe also acknowledge the support of various institutions, including the ARTIC Network, the University of Nebraska, and the Africa Pathogen Genomic Initiative, among others, for their contributions to genomic surveillance efforts in the DRC.
This genome sequencing data is being shared pre-publication and should be regarded as preliminary. Ongoing analyses are expected to provide further insights, and a formal publication detailing these findings is in preparation. Researchers interested in utilizing this data prior to publication are encouraged to contact Prof. Placide Mbala-Kingebeni.
In conclusion, the DRC's latest outbreak of EVD presents significant public health challenges amid other concurrent outbreaks. Ongoing genomic surveillance and collaborative research efforts are crucial for understanding and controlling the spread of Ebola and other infectious diseases in the region.